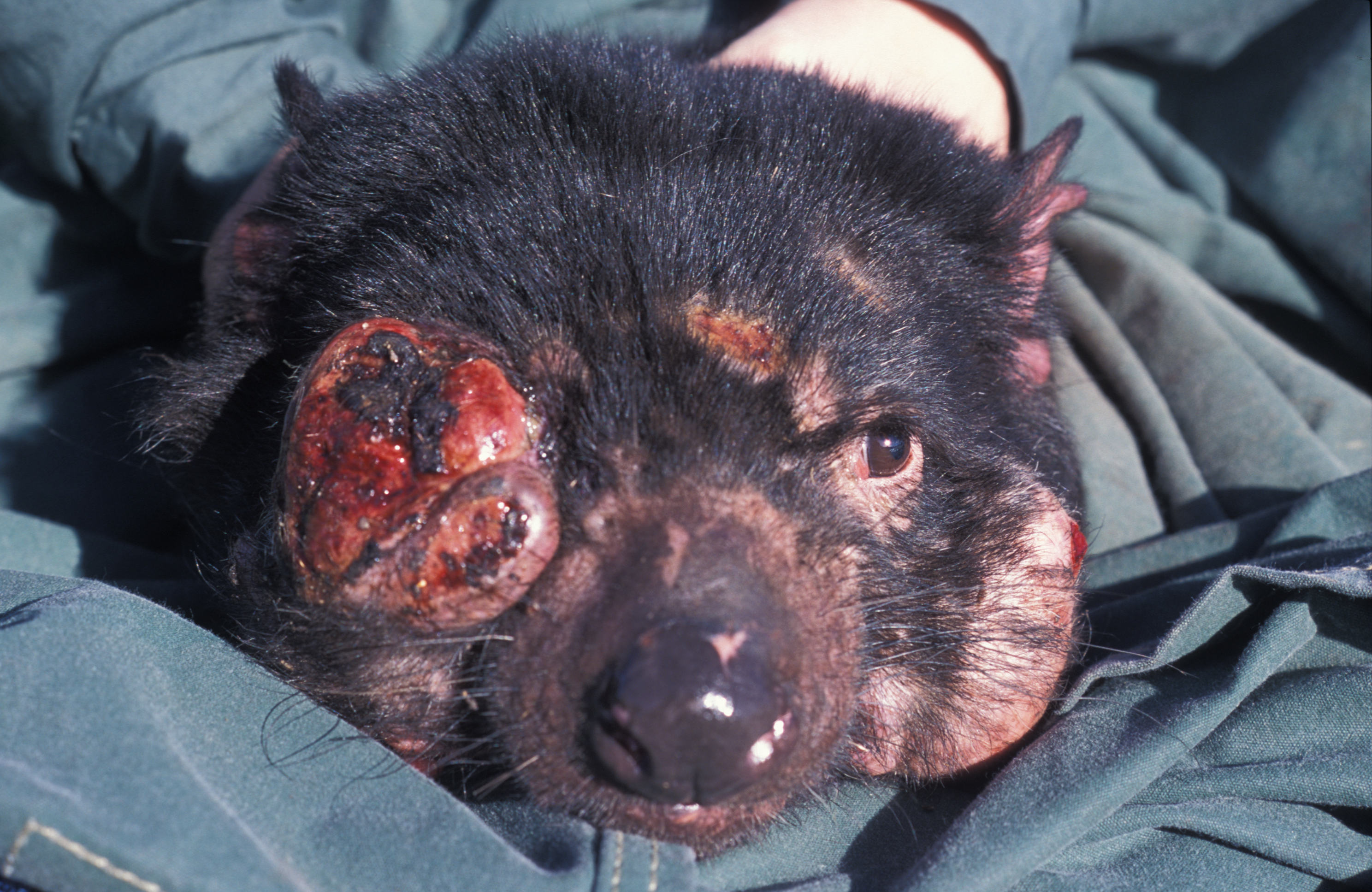
ABOVE: A Tasmanian devil (Sarcophilus harrisii)
MENNA JONES
On the whole, the 20th century was pretty rough for Tasmanian devils (Sarcophilus harrisii). Just when their numbers seemed to finally be recovering after more than a century of poisoning and trapping, a strange, deadly disease emerged: the contagious cancer known as devil facial tumor disease (DFTD). Infected devils grow large, disfiguring tumors, especially on their faces—hence the name—and the vast majority die. In fact, within five years of DFTD’s first appearance in 1996, populations hit by the disease had declined by around 80 percent, and scientists feared the cancer would wipe the devils out in a matter of decades.

These days, the disease continues to kill the world’s largest marsupial carnivore, and populations remain concerningly small. But, according to experts such as Menna Jones, a...
See “The Devil’s Details”
The research, led by University of Idaho’s Paul Hohenlohe and then–graduate student Amanda Stahlke, is the largest genetic study of Tasmanian devils to date, and it uncovered hundreds of genomic regions changing in response to the disease—regions that do not appear to have been under selection previously in the animals’ history.
Billie Lazenby, a wildlife biologist with the Department of Primary Industries, Parks, Water and Environment (DPIPWE) in Tasmania who was not involved in the work, says the findings are “encouraging news” because they build on earlier evidence from Hohenlohe and his collaborators that Tasmanian devils can evolve resistance to DFTD. “If we’re getting selection on these genes . . . it’s indicative that we could see some level of population recovery,” she notes. “It’s also great in the sense that it gives us some context for understanding devil facial tumor disease and its impact through time.”
DFTD resistance: A novel, genome-wide adaptation
Stahlke, now a computational biologist for the US Department of Agriculture’s Agricultural Research Service, says she was drawn to the Tasmanian devil project in Hohenlohe’s lab because of her broader interest in using genomic approaches for conservation. Identifying the genetic basis of any emerging resistance to DFTD could bolster ongoing efforts of DPIPWE and its partners in the Save the Tasmanian Devil Program, she says, as genetic assays could be used to track the spread of resistance in the wild or even screen captive-bred individuals prior to release.
“An evolutionary response to natural selection means, oftentimes, small changes in the frequencies of different genotypes or different genetic variants in a population, and so to be able to detect those changes requires having a lot of individuals,” says Hohenlohe. To get a large enough dataset in their search for resistance variants, the researchers turned to Jones and her field program in Tasmania that’s been studying devils for more than two decades. “We’ve got one of the best sample data sets and genetic sample sets for any emerging infectious disease in wildlife globally,” says Jones.
In all, the team sequenced roughly 16,000 single nucleotide polymorphisms (SNPs) from about 2,500 individual Tasmanian devils in six populations across Tasmania. Some of the loci were chosen because of their previous association with DFTD, cancer, or immunity in general; others, which were at least 20 kilobases away from other loci, were added to ensure a greater coverage of the genome (whole genome sequencing for that many individuals would have been too expensive, Stahlke notes). The team then analyzed how the frequencies of these SNPs changed in each population over time using several different analyses to account for fluctuating population sizes and other potentially confounding effects.
Overall, they found evidence for selection in hundreds of loci scattered across the animals’ genome, which shows that “adaptation to DFTD is a really complicated, organism-wide response,” says Stahlke. Many of these loci were in immune-related genes, including ones previously associated with resistance and/or tumor remission in devils.
There were notable exceptions. Some of the genes have roles in the central nervous system, which could indicate behavioral evolution, or could have something to do with the fact that DFTD initially arose from Schwann cells in the central nervous system before becoming transmissible. The analyses also turned up selection in a gene that, in humans, is associated with ear formation, Stahlke notes; exactly how it might be involved in DFTD response is unclear, but discovering that link could provide novel insights into the animals’ disease response.
Other analyses in the paper suggested this kind of disease is new to the devils. “I was pretty convinced of this hypothesis that devils had seen transmissible cancer previously,” and therefore would have had a remnant of past adaptation to such a disease in their genome, Stahlke says. This idea arose because devils are afflicted with two different transmissible cancers, and such diseases are considered extremely rare. To find out, the team compared the devil’s genome to those of other marsupials to identify genes that rapidly evolved since the devils’ split from their kin, which would suggest historical selection. While they did find a number of genes that fit the bill, there was “very little overlap” with the genes evolving since DFTD cropped up, she says, which suggests contagious cancers are a new challenge to the animals’ survival; presumably, the genes that rapidly evolved previously were in response to other selective pressures.
An evolving situation
Because of the dire predictions for the species early on, a large network of Tasmanian devil captive breeding programs was established in the early 2000s. In theory, this captive population could supplement losses to genetic diversity that occurred because of the epidemic, or could even be used to reintroduce the animals if the disease wiped out their wild kin completely. Now, Jones says, those captive animals could end up harming wild populations.
See “How to Reintroduce a Long-Lost Species”
“They’re getting increasingly out of step with the natural evolution occurring in the wild,” she explains, and because of that, adding them to the wild gene pool could “water down” the emerging disease resistance and ultimately end up fueling a new epidemic of DFTD.
Adaptation to DFTD is a really complicated, organism-wide response.
—Amanda Stahlke, USDA Agricultural Research Service
Lazenby notes that DPIPWE currently has four sites where captive animals are being released to augment local genetic diversity. The Save the Tasmanian Devil team is closely observing the released animals and the populations as a whole to determine exactly what effects the releases might have on those populations, and “this paper really highlights the importance of making sure that we do that,” she says. “It’s really important that, in our efforts to ensure that we maintain as much genetic diversity as we can, we don’t dilute this potential for evolution of tolerance or resistance.”
A bountiful genetic resource
Although compiled to examine selection and provide a means for tracking resistance in devils, the data set the authors compiled could prove useful in many ways, such as investigating whether the disease prompts an increase in inbreeding, notes Anna Brüniche-Olsen, a conservation geneticist at the University of Copenhagen who investigated selection in Tasmanian devils during her doctoral training (Jones was one of her PhD advisors).
And DFTD isn’t the only contagious cancer the animals face. The second transmissible tumor, DFT2, was first spotted in 2014. It’s currently limited in range, but is expected to spread much like DFTD did. “It will be interesting, in the future, to try to compare [what happens genomically after] DFTD and this DFT2,” Brüniche-Olsen says, something “this data set could be excellent for.”
Brüniche-Olsen says she’d love to see the sequencing expanded to whole genomes. The 16,000 loci used represent a small portion of the animal’s genome, she says, and she “wouldn’t be surprised” if there are genomic regions under selection that were missed by the study. She’d also like to see comparisons with other species that have contagious cancers to see if any of the same genes show evidence for selection, as it’s possible other changing aspects of the devils’ environment are confounding the results. “That would be more like an independent validation” of the genes’ importance in resistance to DFTD.
See “Second Contagious Cancer Found in Tasmanian Devils”
Lazenby notes that the findings could aid research into potential therapeutics, from helping determine which human cancer drugs have the greatest potential for treating devils to guiding vaccine development efforts—but researchers will need to connect the selected-for genes to functional changes first. Also, linking genes to physiology more directly could reveal any potential downsides or tradeoffs involved. “By having this strong selection, does that make devils susceptible to anything else moving into the future?” asks Lazenby. “I think it’s something which would be good to consider.”
In the meantime, Jones says, the results indicate that the devil will recover all on its own. “It’s got enough standing genetic variation to adapt, so we don’t need to wait for new mutations” she says. “The devil is going to live with [DFTD]. It’s not going to become extinct.”
Interested in reading more?







