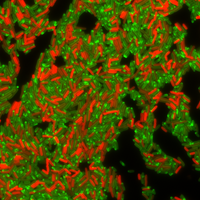
ABOVE: Some bacterial cells produce a drug-resistant protein (labeled in red). Drug-sensitive cells swamped in the antibiotic tetracycline are visible in green.
CHRISTIAN LESTERLIN & SOPHIE NOLIVOS, UNIVERSITY OF LYON
Escherichia coli is capable of synthesizing drug-resistant proteins even in the presence of antibiotics designed to cripple cell growth. That’s the finding by a group of French researchers reporting today (May 23) in Science. They also discovered how the bacteria manage this feat: a well-conserved membrane pump shuttles antibiotics out of the cell—just long enough to buy the cells time to receive DNA from neighbor cells that codes for a drug-resistant protein.
“This is a key discovery,” microbiologist Manuel Varela of Eastern New Mexico University who wasn’t involved in the study says to The Scientist in an email. “This finding will help explain how bacteria manage to spread antimicrobial resistance as they encounter toxic levels of antibiotic.”
The...
Using E. coli’s habitual sharing of antibiotic resistance genes as a case study, they watched as bacteria passed around DNA encoding the TetA protein—a pump that makes cells resistant to tetracycline by shunting it out of the cell. Shortly after, they observed plasmid DNA arriving in non-resistant cells, and some time later, red fluorescent spots appearing in the membranes of recipient cells, indicating the TetA protein was translated and the cells proved resistant to tetracycline.
The antibiotic, which is commonly used in livestock, but sometimes also to treat people for pneumonia, respiratory tract infections, and other conditions, ordinarily stunts the growth of bacteria that don’t have TetA, but numerous bacteria strains are becoming resistant through the adoption of such mechanisms. Tetracycline wasn’t present for this initial experiment, so to see how this process is influenced by the drug itself, the researchers exposed the cells to high concentrations of tetracycline and once again put them under the microscope.
As expected, they observed plasmid DNA arriving in new, non-resistant cells. This was expected, because tetracycline does nothing to hinder that process. Instead, it’s designed to stall protein production. And surprisingly, the researchers saw the red fluorescence appearing in the some of the new recipient cells that didn’t previously have the TetA protein: evidently, they were still able to synthesize proteins, including TetA, despite being exposed to tetracycline. “We spent many, many weeks just confirming this result, which was very counterintuitive, and we had a hard time being convinced that it was actually happening,” recalls Lesterlin.

The team made an educated guess as to why the cells were capable of this: many bacterial membranes are known to harbor a multidrug efflux pump known as AcrAB-TolC, which is capable of shuttling a wide range of antibiotics out of cells, and the scientists figured that it was getting tetracycline out of the cell before it could stop protein synthesis and cell growth. To test that idea, the researchers engineered several mutants with a genetic mutation in one of the genes that encodes the different proteins that make up the pump.
They found that the mutants, although they received the plasmid bearing the genetic code for TetA from neighboring cells, weren’t capable of synthesizing TetA protein. Without the functional efflux pump, the mutants can’t shuttle the tetracycline out of the cells. As levels of the antibiotic surged inside the cells, they could no longer make proteins or grow.
When functional, the AcrAB-TolC pump buys the bacteria time by keeping antibiotic concentrations just low enough for the cells to synthesize the resistance proteins encoded in the plasmid DNA, according to the researchers. In this case, it allows for the production of the TetA protein, which then shunts more tetracycline out of the cell. Ultimately, bacteria can become resistant while still under the influence of antibiotics. As Lesterlin puts it, “better news for bacteria than for human health.”
“The multidrug efflux pump AcrAB-TolC has been known in the field for quite some time,” notes Anushree Chatterjee, a chemical engineer and microbiologist at the University of Colorado Boulder who wasn’t involved in the study. But the fact that it helps bacteria acquire drug resistance while they are simultaneously exposed to antibiotics is news, she says. “It’s always fascinating to see how there’s so many things bugs can do.”
The findings are widely relevant, she says—for one, because AcrAB-TolC is so broadly conserved across bacteria, and also because the mechanism is not limited to tetracycline. Lesterlin and his colleagues demonstrated that the pump also allows bacteria to produce drug-resistant proteins in the presence of other antibiotics designed to stifle gene expression, such as the translation-inhibiting chloramphenicol, and the transcription-inhibiting rifampicin. This mechanism is relevant for so-called bacteriostatic antibiotics, which don’t kill but only stifle bacterial growth, Lesterlin adds. He doubts it will work for bacteriolytic antibiotics, which destroy bacteria outright before they can become resistant.
Both Chatterjee and Varela find the new study thorough and its findings robust, Varela being particularly impressed by the technique the team developed to visualize the transfer of plasmid DNA between cells while watching TetA protein synthesis at the same time.
“The authors have [also] shed light on identifying key bacterial machinery that could serve as new targets for developing new anti-bacterial agents,” Varela adds in an email. For instance, one could build antibiotics by targeting the AcrAB-TolC pump—an approach some labs are already working on. Alternatively, one could target genes that regulate its production—an angle that appeals to Chatterjee. Traditional approaches of designing antibiotics have largely relied on small molecules that target specific proteins, many of which bacteria have seen for many years and ultimately select for more resistance mechanisms.
“We need to look at non-traditional pathways,” Chatterjee says. “What are the regulatory mechanisms that allow cells to navigate these stressful situations? I think targeting those processes seem to be a pathway towards building smarter therapies that can hopefully thwart resistance from the very beginning.”
S. Nolivos et al., “Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer,” Science, doi:10.1126/science.aax6620, 2019.
Interested in reading more?