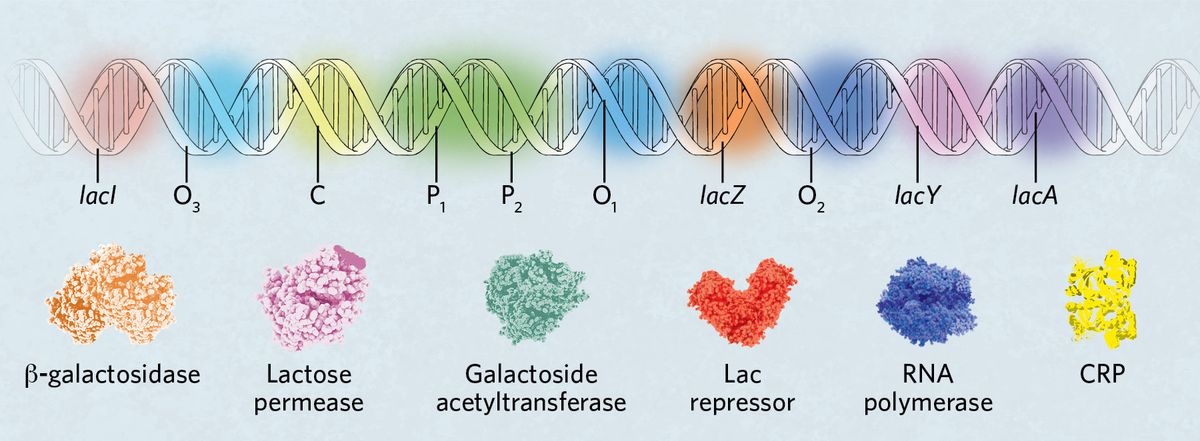
The genes and important sequences for the regulation of the lac operon are organized such that several mediators can fine tune expression of the three genes in the operon itself—lacZ, lacY, and lacA. The gene lacI, which encodes the lac repressor, LacI, sits outside the operon and is constitutively expressed. A cyclic adenosine monophosphate (cAMP) receptor protein (CRP) binding site (C) sits in front of the promoter region where there are two potential RNA polymerase binding sites (P1 and P2). CRP controls the binding of RNA polymerase between P1 and P2 by binding C. Finally, three operator regions, O1, O2, and O3, are spaced through the DNA region to modulate LacI binding and overall operon repression.
Leaky Repression
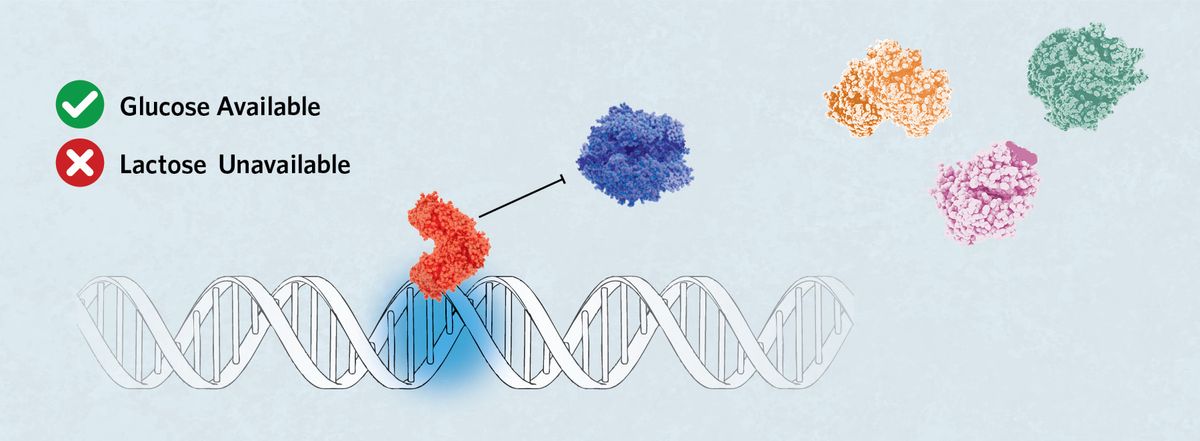
When glucose, but not lactose, is available in the cell, the LacI binds to the O1 sequence, preventing RNA polymerase from binding to the promoter region. However, due to protein kinetics, a small amount of the lac operon genes can be expressed if the polymerase binds when one repressor releases the DNA before another binds.
Strong Activation
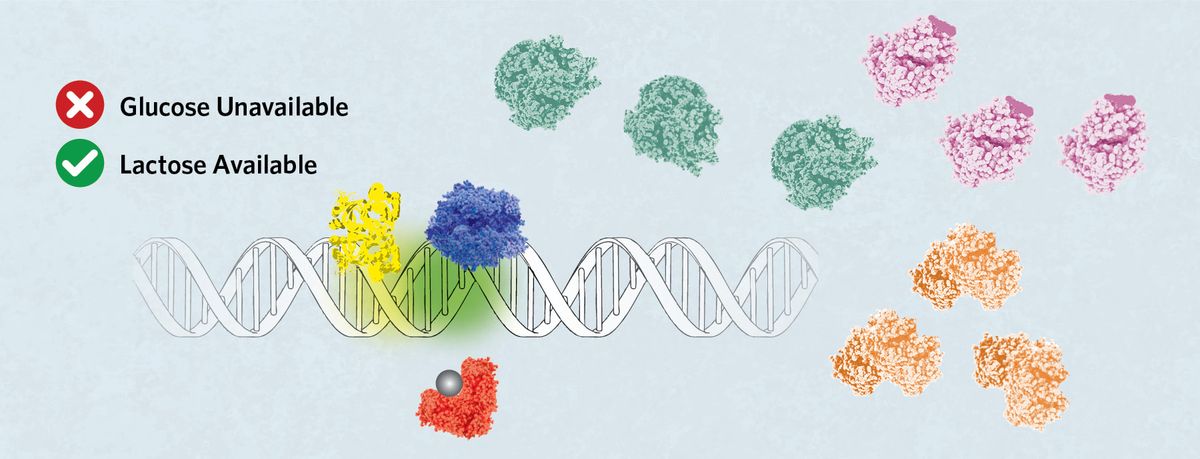
If glucose becomes unavailable but lactose is present, then allolactose (grey sphere) binds LacI, releasing it from the operator. cAMP, produced in the absence of glucose, attaches to CRP, prompting its binding to the C site. This directs the RNA polymerase to sit on the P1 site, which promotes robust expression of the lac operon to facilitate lactose digestion.
Tight Repression
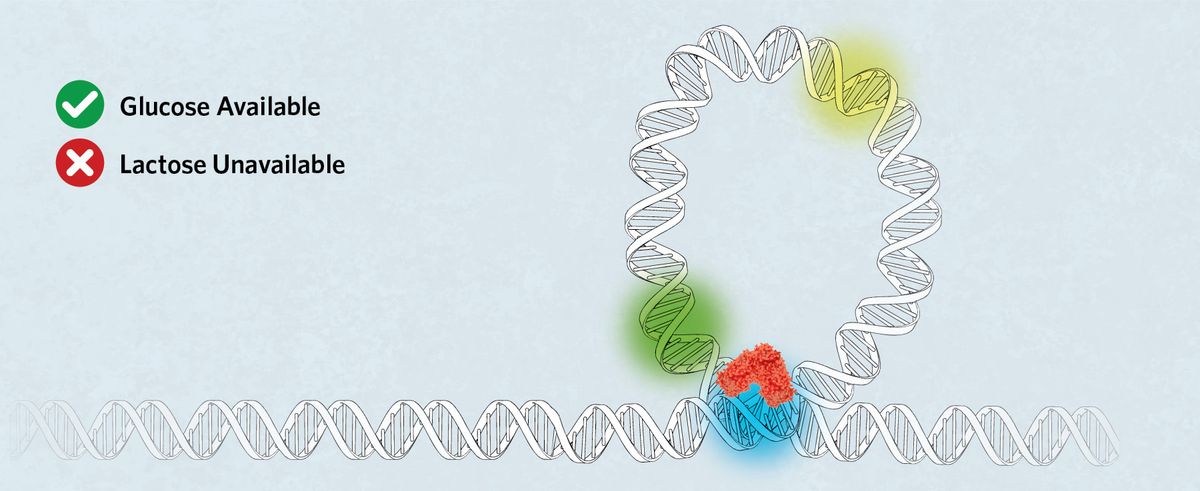
The repression of the lac operon in the absence of lactose can be improved through DNA looping, in which LacI binds to O1 and a second operator sequence, either O2 or O3. This increases the local concentration of LacI, reducing transient expression that occurs when only free-binding lac repressor is available.1
Weak Activation
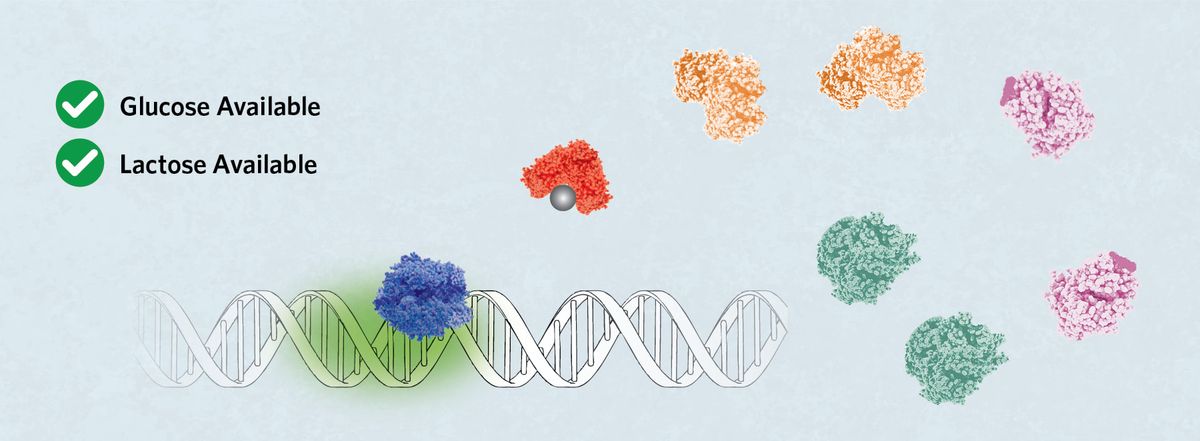
When glucose and lactose are both available, allolactose releases LacI from the operator, allowing binding of the RNA polymerase. However, in the absence of cAMP to permit CRP binding, the polymerase binds either P1 or P2 and does not remain on the DNA as effectively, leading to low expression of the lac operon genes.
Read the full story.
- Vilar JMG, Leibler S. DNA looping and physical constraints on transcription regulation. J Mol Biol. 2003;331(5):981-989.
