ABOVE: The 2024 Top 10 Innovation winners include technologies involving advanced cell sorting and analysis, artificial intelligence, omics, CRISPR, and medical imaging and monitoring. The Scientist
The Scientist’s Top 10 Innovations contest is back. This year we took nominations for cutting-edge products launched between January 2023 and July 2024. The results showcased emerging trends in basic, translational, and clinical research, and beyond.
From fine-tuned updates to brand-new products set to shake up the ways scientists conduct research and improve how clinicians treat their patients, the winner’s circle includes established companies and new startups. Looking at our top products, it is clear that advanced cell sorting and analysis technologies made a big splash this year. Products that provide a spatial spin on cell and tissue analysis, often with the help of artificial intelligence and machine learning, captured our judges’ attention. In addition, omics advances, new technologies for medical imaging and monitoring, and better ways to model disease and detect biomarkers took top ranks. Finally, a teaching tool educating the next generation of scientists rounds out our top innovations list.
We are happy to announce the new technologies that our panel of independent judges has chosen as this year’s Top 10 Innovations.
REM-I Platform | Deepcell

Many technologies help researchers glean insights into cellular phenotypes, but it is challenging to manually identify subtle cellular morphology differences, and more so to do it in an unbiased and quantitative manner. Deepcell’s REM-I platform for single-cell analysis, launched in April 2024, provides critical insights into cellular heterogeneity and functionality by combining artificial intelligence (AI)-based technology and high-throughput imaging.
“REM-I is really the first platform that brings AI capability with high resolution, bias-free, brightfield imaging and physical action as part of sorting into one platform,” explained Maddison Masaeli, Deepcell co-founder and chief executive officer. REM-I uses advanced microfluidics and optics to capture high-resolution brightfield images of each cell in a sample. The platform combines self-supervised learning and morphometrics (computer vision) to collect morphological information and clusters cells in real time based on biophysical properties.
REM-I enables researchers to rapidly view, compare, and sort morphologically distinct cell populations, capturing cells that may otherwise be missed through manual methods. Clinical bioinformatician Peter van der Spek at Erasmus Medical Center, who is on Deepcell’s scientific advisory board, envisions opportunities to improve pathology analyses based on his experience using REM-I for several translational research projects, including investigations into kidney transplant rejection. “We saw at the morphology level, cells that have never been observed by our pathologists. That is very fascinating because you're starting to see new shapes, new states of cells which actually open up the biology,” he explained.
According to Masaeli, Deepcell offers different pricing models for REM-I, which is currently used in research only and is commercially available.
MARDIS: This technology seems to be the new generation of microscopy because it can image and characterize single cells based on AI-training of morphology, rather than relying on human eye-brain characterization.

CosmoSort | Meteor Biotech
Most bulk cell sorters remove cells from their spatial context. Meteor Biotech’s CosmoSort cell sorter retains the original spatial information while isolating stained target cells based on structural and cellular imaging data.
According to Sumin Lee, Meteor Biotech’s chief technology officer, engineers at Seoul National University initially developed the CosmoSort technology based on collaborations with biomedical researchers looking to better answer clinical questions. “We thought, what if we could directly sort out the cells from pathology tissues, so that we can deeply analyze those cells?” said Sumin Lee.
CosmoSort’s spatially-resolved laser activated cell sorting technology uses near-infrared laser pulses to rapidly release targeted cells within slide-mounted specimens. The instrument preserves the collected cells’ natural states and spatial information, while sorting them into retrieval vessels. Scientists can then perform various downstream omics analyses with the isolated cells to gain spatial insights into cellular diversity and disease pathology.
Using this innovation, scientists can derive detailed data about an individual’s tumor microenvironment. Han-Byoel Lee, a clinician specializing in breast cancer surgery and research at Seoul National University, was involved in several projects assessing tumor tissue using earlier versions of the CosmoSort technology. In one study, after isolating regions of interest from breast cancer tissue, “we performed whole transcriptome and epi-transcriptome analyses on the cells,” said Han-Byoel Lee.1 “We were able to identify adenosine-to-inosine editomes that may be related to resistance of therapy in triple-negative breast cancer.”
Meteor Biotech launched its first CosmoSort instrument in March 2024, and their scientists are currently working on updating the machine with integrated fluorescence microscopes and AI-driven features.
HOCKBERGER: [The instrument] facilitates transcriptome and epi-transcriptomic insights that shed light on cellular functions in situ. This technology is a powerful addition to the single-cell multiomics toolbox.

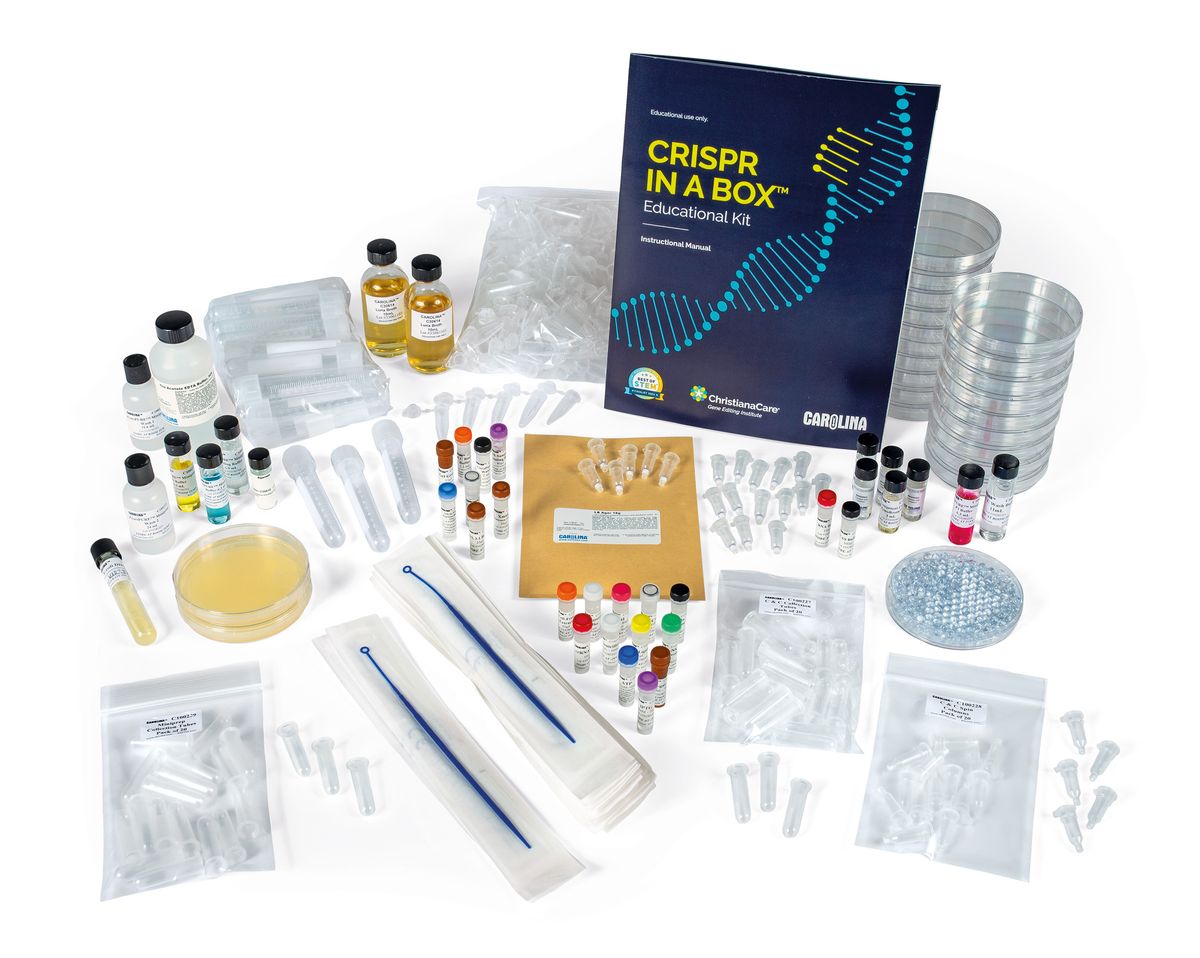
CRISPR in a Box™ | ChristianaCare® Gene Editing Institute

The entertainment industry often misrepresents CRISPR genome editing, leaving the public confused about how this technology works. However, the emergence of gene therapies has sparked excitement in high school and college students, inspiring them to learn more. As a result, the ChristianaCare® Gene Editing Institute developed the CRISPR in a Box™ educational kit and partnered with Carolina® Biological Supply Company to manufacture and distribute the current version, which they released in March 2024.
“[The kit offers] an opportunity to really expand the students’ understanding through hands-on, active learning,” said Philip Kudish, a science instructor at Delaware Technical Community College, who received the kit for beta testing. “One of the things that makes CRISPR in a Box so versatile is that the CRISPR gene editing step is done in vitro.” Typically, CRISPR cleavage and gene repair processes occur within target cells. However, the gene editing step in CRISPR in a Box does not require cells, which allows educators without access to cell culture equipment to still perform this portion of the exercise with their class.
“We are hoping that [CRISPR in a Box] is going to demystify gene editing technology,” said Amanda Hewes, one of the inventors of the kit and the educational program manager at the ChristianaCare Gene Editing Institute. “[Using this kit], we can teach the younger generations about what CRISPR and gene editing could do, whether it is in healthcare, agriculture, or any other fields,…and hopefully train some of the next generation of scientists, teachers, and professionals that are going to get into the [gene editing] field.”
HOCKBERGER: The kit includes print materials and hands-on exercises for high school and college students interested in learning the basics of CRISPR gene editing.
Tensor Field Mapping | Q Bio

Magnetic resonance imaging (MRI) is a staple of medical diagnostics, but its power has been hampered by data variability and the technique’s qualitative nature. In May 2024, Q Bio Inc. unveiled Tensor Field Mapping (TFM), a new MRI technology that provides quantitative imaging data that is harmonized across instruments and operators. TFM represents a “quantum leap forward,” said Q Bio founder, chief technology officer, and chief executive officer Jeff Kaditz in a statement. Kaditz further stated that TFM makes “multi-parametric quantification of tissues reproducible across machines, which is key to [artificial intelligence] in MRI and personalized medicine.”
TFM technology will debut in Q Bio’s Mark I full-body scanner, an instrument under development as of October 2024 that was specifically designed to take advantage of what TFM can accomplish. “Quantified MRIs have been anticipated for over 25 years,” explained Thomas Witzel, vice president of imaging at Q Bio, in a press release. “By modeling the entire imaging process without idealizing assumptions about the hardware, we're not only enhancing the quality of MRI data but also paving the way for developing new MRI equipment optimized for speed and patient comfort.”
Q Bio is currently looking for partners to test and prove the capabilities of TFM within real-world health systems. “We look forward to sharing our collaborations with healthcare providers and researchers to bring the benefits of TFM to patients around the world,” said chief operating officer Clarissa Shen in a statement.
MARDIS: The advanced capabilities of this new MRI technology can eliminate variability between different MRI instruments, making quantitative measures of tissues conducive to machine learning or AI-based improvements.
UG100™ | Ultima Genomics
DNA sequencing has evolved to support the analysis of large quantities of genetic material and the production of vast amounts of data. Researchers seek solutions for workflow bottlenecks, and one of the first places they turn to is sequencer technology.
February 2024 marked the full commercial release of Ultima Genomics’ UG100™ high-throughput sequencer, which uses cutting-edge technology to improve sequencing speed and efficiency. “We have brought to market a high-throughput, high-quality sequencer that sequences smaller batches at a lower price point that any other sequencer available,” said Jay Therrien, Ultima Genomics’ chief commercial officer. The company prides itself on the UG100’s ability to reduce costs to 100 USD per genome.
Leading researcher institutes globally already use the platform. Cheaper and faster sequencing supports data generation and research discovery, enabling information extraction from more cells, tissue types, samples, time points, and treatments. “The more that we lower the price of generating data, the more can be done. We see that take hold in a variety of market segments,” Therrien said.
Among the first adopters of the technology were scientists at Macrogen Inc., a company that produces DNA sequence reads for their research customers and the public. Seung-been “Steven” Lee, head of the Technology Strategy Office at Macrogen Inc., describes UG100’s cost-effectiveness, automation, and reliability as a game changer. The platform uses a special spinning silicon wafer to distribute reagents, which speeds up data collection and imaging tremendously. “The UG100 can store up to six wafers, that’s six flow cells, and it automatically starts a new run,” Lee said. “The automation has been working very well, and it really improved the efficiency of our pipeline.”
KAMDAR: The Ultima UG100 system offers a platform for precision and low-cost genomic sequencing, offering unparalleled accuracy and speed to unlock deeper insights into the complexity of life’s code.

Microscoop™ | Syncell

Scientists increasingly turn to microscopy-based spatial biology techniques to detect, localize, and map proteins, but high-throughput or unbiased spatial proteomics screening remains limited by method resolution, sensitivity, and specificity. Syncell’s Microscoop™ brings microscopy-guided proteomic discovery into the automated, large-scale realm.
Microscoop is a new spatial biology platform that leverages AI and advanced image processing, precise photolabeling-based biotinylation, and a streptavidin bead pull down assay for mass spectrometry analysis. It performs automated photolabeling for all fields of view in a sample and can rapidly reveal new proteins at specific subcellular regions, such as protein aggregates in brain tissue or biomarkers in cancer samples. “This technology is probably the first high precision spatial protein purification method,” said Jung-Chi Liao, Syncell’s chief executive officer. “You can pull [down] the proteins in the location you're interested in and identify what they are in a much better, much faster way.”
“What many companies now call spatial proteomics is often candidate-based…Here the technology is quite different, because in this case, it's really discovery-based,” said neurobiologist Wilfried Rossoll from Mayo Clinic, who received early access to the Microscoop system as a winner of Syncell’s Spatial Proteomics Grant Opportunity Award. Rossoll’s research team employed the platform to investigate key neurodegenerative disease markers within pathological protein aggregates in human autopsy brain tissue.
Rossoll also led the initiative to bring a mass spectrometry core facility to the Mayo Clinic in Florida, where one of the first Microscoop systems has been installed in the US. The official launch of the Microscoop system coincides with the 2024 American Society for Cell Biology meeting, a representative for Syncell said via email.
MEAGHER: This is a cool approach to sub-cellular spatial proteomics that combines automated microscopy and targeted photolabeling to enabled unbiased proteomics of selected sub-cellular features.

STARVUE™ | Vizgen (Ultivue)

When performing spatial image analyses of heterogeneous tissues, the amount of data generated can become unmanageable. “If you want to study, for instance, the tumor microenvironment, you need to understand what different cell types are in your tissue, what's the actual battle ongoing there between the immune system and cancer cells,” said Lorenz Rognoni, the spatial image analysis portfolio owner at Vizgen (Ultivue). “If you then multiplex, you have to overcome this massive data challenge.”
To facilitate the exploration of complex tissues, Vizgen offers a range of VUE multiplex immunofluorescence (mIF) biomarker panels, which generate rich mIF images of stained slides. In April 2024, the company rolled out the STARVUE™ Image Data Science Platform to handle this plethora of data.
STARVUE uses pre-trained, tissue-based image analysis applications to analyze mIF images of whole stained slides, providing tissue detection, segmentation, and cell-biomarker classification based on morphology and stain signals. The cloud-based, automated platform can stack whole slide mIF and same-section hematoxylin and eosin (H&E) stain images, and its deep learning models classify cells throughout complex and variable tissues with the help of artificial intelligence.
Scott Daigle, senior director of Translational Medicine at Nimbus Therapeutics, highlighted how scientists at his company leverage the VUE panels and STARVUE to identify oncology targets that modulate the immune system. “We can get majority of our answers just from this one assay…Are those T cells engaging with the cancer cells, are they activated, are they doing as we expected?” said Daigle. “Tissues from clinical samples are extremely precious, and so to be able to multiplex and get that type of data is great.”
KAMDAR: STARVUE empowers researchers with high-definition spatial biology, delivering unprecedented clarity and multiplexing capabilities to unravel complex tissue biology with ease and precision.
Direct RNA Sequencing | Oxford Nanopore Technologies

Transcriptomic analysis is essential for characterizing cells in health and disease. But most RNA sequencing techniques do not directly analyze native RNA molecules and instead sequence cDNAs. The reverse transcription process loses vital information about the RNA molecules including base modifications.
To overcome this limitation, Oxford Nanopore Technologies developed a direct RNA sequencing workflow, which requires a MinION™ nanopore sequencing device, a MinION RNA Flow Cell, and a Direct RNA Sequencing Kit. This system enables researchers to comprehensively examine an RNA molecule’s sequence and detect several post-translational modifications including N6-methyladenosine, pseudouridine, inosine, and 5-methylcytosine.
Oxford Nanopore released the latest version of the flow cell and sequencing kit, known as RNA004, in March 2023. “What we have done with RNA004 over old versions is improved its accuracy, improved its output, and reduced the input material requirement,” said Lakmal Jayasinghe, the senior vice president of R&D Biologics at Oxford Nanopore.
Additionally, the updated direct RNA sequencing system can now analyze short RNAs that are 50 nucleotides or longer. “The ability to read RNA molecules that are otherwise very hard to [read including] non-coding RNA, structured ribosomal RNA, or tRNA…is remarkable and quite empowering,” said Miten Jain, a nanopore sequencing expert at Northeastern University, who received the system for beta testing. “[With direct RNA sequencing], we can start looking at cancer cells, neurodegeneration, or pancreatic metabolism-associated models and get a better and more complete picture of the landscape of RNA, not just expression, but also modifications.”
MEAGHER: I’m excited to see the technology mature because I think this will open new application areas that can’t be addressed by traditional RNA-seq methods that start by reverse-transcribing RNA to cDNA prior to sequencing.

Proteograph XT Assay Kit | Seer

Biotech company Seer released the Proteograph™ Product Suite in January 2022, giving researchers a way to study unknown and low-abundance proteins. However, they soon received requests for more throughput and even more depth from scientists looking to push the envelope, recalled David Horn, chief financial officer for Seer. The Proteograph XT Assay Kit, launched in June 2023, is Seer’s answer. “The original product could process 16 samples per eight-hour run,” explained Horn. “With the XT, now you can do up to 80. This allows for large-scale studies, which we have seen increasing interest in.” Horn said that this improved throughput enables studies looking at up to 50,000 samples, and he believes that these large-scale studies will enable a better understanding of biology.
Biochemist Joshua Coon has seen this first-hand. When the COVID-19 pandemic started in spring 2020, Coon and his team performed proteomic analyses on plasma samples. “This was before the Proteograph, and we were able detect about 300-350 proteins per sample.” Since then, the project has moved to studying long COVID: “This summer, with the Proteograph, we are now getting 6,500–7,000 proteins per sample. We are also getting both the markers that we saw in the first study and new markers that have clear associations. We may be able to separate markers for long COVID from markers for acute COVID, so I am super excited,” said Coon.
MEAGHER:This is a clever approach to address problems of bias and dynamic range in proteomics, by using different sets of nanoparticles to selectively sample for different representations of the proteome, and thereby gain better depth of coverage of the proteome.

PhysioMimix® Single-Organ Higher Throughput (HT) System | CN Bio

Accurately reproducing the unique microphysiology of human organs in culture is a prerequisite for generating reliable preclinical data that mimics human physiology and improves translatability. This is especially true for scientists engaged in drug discovery and development research.
Launched in February 2023, CN Bio’s PhysioMimix® Single-Organ Higher Throughput (HT) System is a perfused, organ-on-a-chip platform that addresses the need for large-scale studies to generate robust, human relevant, predictive, and cost-effective preclinical data. “This system has a total capacity of 144 chips, which can be run simultaneously,” said Paul Brooks, chief executive officer of CN Bio. This is particularly useful for toxicology and efficacy studies, where informed decisions about data sufficiency mitigate drug failure in future clinical trials. Brooks highlighted configurability as one of the platform’s key advantages.
The platform bridges traditional cell culture and the complexity of animal studies, allowing researchers to minimize the number of animal models while using a human cell-based system that remains faithful to the physiology of human tissues and organs. Alysha Bray, a scientist at CN Bio, believes that this technology, plus their Liver-48 plate for 3D culture, simplifies the use of organ-on-a-chip approaches in laboratories, increasing research capacity and throughput and reducing costs by using fewer primary cells. “Despite the smaller size of [each] well and tissue, we don’t have to compromise on quality of the data produced,” Bray said. “Using the Liver-48 plate is straightforward, and the fact you can use a multi-channel pipette for changing media and taking samples is a real time saver. Every moment counts in the lab, and this higher-throughput system allows us to do more whilst using less.”
HOCKBERGER: CN Bio’s new higher-throughput organ-on-a-chip technology is poised to revolutionize drug discovery, toxicology, and modeling of treatments for human liver diseases.

REMI™ Remote EEG Monitoring System | Epitel™
The seizure disorder epilepsy affects roughly 3.4 million people in the US. Beyond epilepsy, one in 10 people will have a seizure in their lifetime, underscoring the importance of seizure detection. However, access to neurologists and electroencephalogram (EEG) monitors that record brain activity remains a gap in the healthcare system. “Today you could walk into your neighborhood clinic and walk out with a wireless halter monitor for your heart, but your chances of getting an EEG are slim to none. In fact, three quarters of all clinics lack the ability to get an EEG,” said Epitel™ chief executive officer and chief technology officer Mark Lehmkuhle.
Epitel's REMI™ is the first wireless wearable EEG system, enhancing epilepsy management through advanced remote monitoring. It allows healthcare professionals to track seizure activity in real time and outside of the clinic. The system includes four adhesive sensors that contain conductive hydrogel, which wirelessly transmit signals to a dedicated smartphone. Machine learning analyzes data stored in the cloud and sends readouts to healthcare providers, making it easier to identify and monitor seizure activity. REMI is FDA-cleared for home use for up to 30 days by prescription.
“Access to neurology and neurologists is limited and will be continually limited in the future. Technology like this can help bridge that gap and make it so there's greater access,” said neurologist Steven Benedict at Northern Ohio Medical Specialists, who first used REMI in a clinical outpatient setting during beta testing. “Just having access, for the patients and for the providers, and being able to answer clinical questions easily, is a tremendous benefit.”
KAMDAR: Epitel’s REMI revolutionizes remote EEG monitoring, offering real-time, accessible insights into brain activity that empower clinicians to make faster, more informed decisions for better patient outcomes.

VisionSort™ | ThinkCyte

Launched in the summer of 2023, ThinkCyte’s VisionSort™ adds advanced optics, microfluidics, high-resolution morphological profiling, and AI to conventional cytometry.
Traditional flow cytometry requires fluorescent labels, which can alter cells’ native states and cannot capture cell morphology at high resolution. VisionSort is a multi-laser flow cytometer that uses Ghost Cytometry® technology, a cellular fingerprinting method that combines morphological profiling, fluorescence, and AI to define and sort cell populations label-free. With this instrument, researchers can isolate specific cell populations with high viability using gentle microfluidics, screen for new drugs, and identify novel cell populations in disease.
Since its launch, researchers have used VisionSort for cell therapy development, drug discovery, and disease profiling. “Whether it’s isolating truly untouched and phenotypically defined cells, performing CRISPR-based phenotypic screens by flow cytometry, or identifying novel cell populations in disease, VisionSort gives researchers the ability to do things they couldn’t do before,” said Willem Westra, vice president of Business Development and Marketing at Thinkcyte, in an email.
Scientists at VCCT Inc., a regenerative medicine company that develops cell therapies for retinal degeneration, began using VisionSort in early 2024. “A critical element of our research and development pipeline requires analysis of induced pluripotent stem cell-derived retinal cells, a longstanding challenge due to the lack of specific surface markers, reporters, or chemical labels,” said Masayo Takahashi, president of VCCT Inc., in an email. “This approach allows our researchers to enrich live, phenotypically-defined cells and use them in downstream steps."
MARDIS: This technology couples the innovation of AI-driven cell sorting that is trained on 3D morphology to identify cells, coupled with conventional cell surface-labeling using fluorescent antibodies.

THE JUDGES

Philip Hockberger is internationally recognized for his leadership role in shared research facilities and for his efforts to create a new career path for core scientists. He is a member of the Executive Board of BioImaging, North America and the National Science Foundation’s National Visiting Committee. He was a professor of neuroscience at Northwestern University for 36 years. He has been co-chair of Euro-BioImaging and a member of the Federation of American Societies for Experimental Biology’s Task Force on Shared Research Resources.

Kim Kamdar is a managing partner at Medical Excellence Capital, a healthcare-focused venture fund creating and investing in biopharmaceutical and diagnostic companies. She began her career as a scientist and pursued drug discovery research at Novartis/Syngenta for nine years.

Elaine Mardis is the co-executive director of the Steve and Cindy Rasmussen Institute for Genomic Medicine at Nationwide Children’s Hospital. She holds the Rasmussen Nationwide Foundation Endowed Chair in Genomic Medicine and is a professor of pediatrics at The Ohio State University College of Medicine. Mardis is an expert in cancer genomics and immunogenomics, involved in the clinical profiling of pediatric cancer DNA and RNA to identify somatic and germline contributors to cancer onset and to characterize the tumor immune microenvironment.

Robert Meagher is a chemical engineer by training. His research interests include microfluidics, bioanalytical chemistry, and molecular diagnostics for infectious disease. He is the manager of the Systems Biology Department at Sandia National Laboratories, which performs fundamental research in topics including microbiology, virology, emerging infectious disease, biodefense, and bioenergy.
The statements expressed represent the personal views of Robert Meagher and do not reflect the position of Sandia National Laboratories, National Technology & Engineering Solutions of Sandia, LLC, the National Nuclear Security Administration or the Department of Energy.
- Lee AC, et al. Spatial epitranscriptomics reveals A-to-I editome specific to cancer stem cell microniches. Nat Commun. 2022;13(1):2540.
Membership Open House!
Enjoy OPEN access to Premium Content for a limited timeInterested in exclusive access to more premium content?






